Next-Morning PCR Results for Everyday Infections
HealthTrackRx PCR test results allow providers to diagnose and target treatment for infections by the next morning. Our RT-PCR technology targets the most relevant viral, bacterial, and fungal pathogens across a full menu of everyday infections – UTI, STI, GI, respiratory, chronic wounds, and more.

Next-Morning PCR Results for Everyday Infections
HealthTrackRx PCR test results allow providers to diagnose and target treatment for infections by the next morning. Our RT-PCR technology targets the most relevant viral, bacterial, and fungal pathogens across a full menu of everyday infections – UTI, STI, GI, respiratory, chronic wounds, and more.
Years of Lab Testing
Employees
Patients Served
MDs, PHDs, MS
Antimicrobial Resistance
Rising Healthcare Costs
The cost of Antimicrobial Resistant infections annually has been estimated at $21-34 billion in the US healthcare system alone, which includes more than 8 million additional days in the hospital2. It is estimated that by 2050, AMR may lead to $100 trillion in world GDP loss3. HealthTrackRx is addressing this public health crisis head on through developing advancements in PCR technology and implementing innovative solutions for our clients, that help you treat patients fast, with accuracy.
Advanced Molecular
Diagnostic Solutions
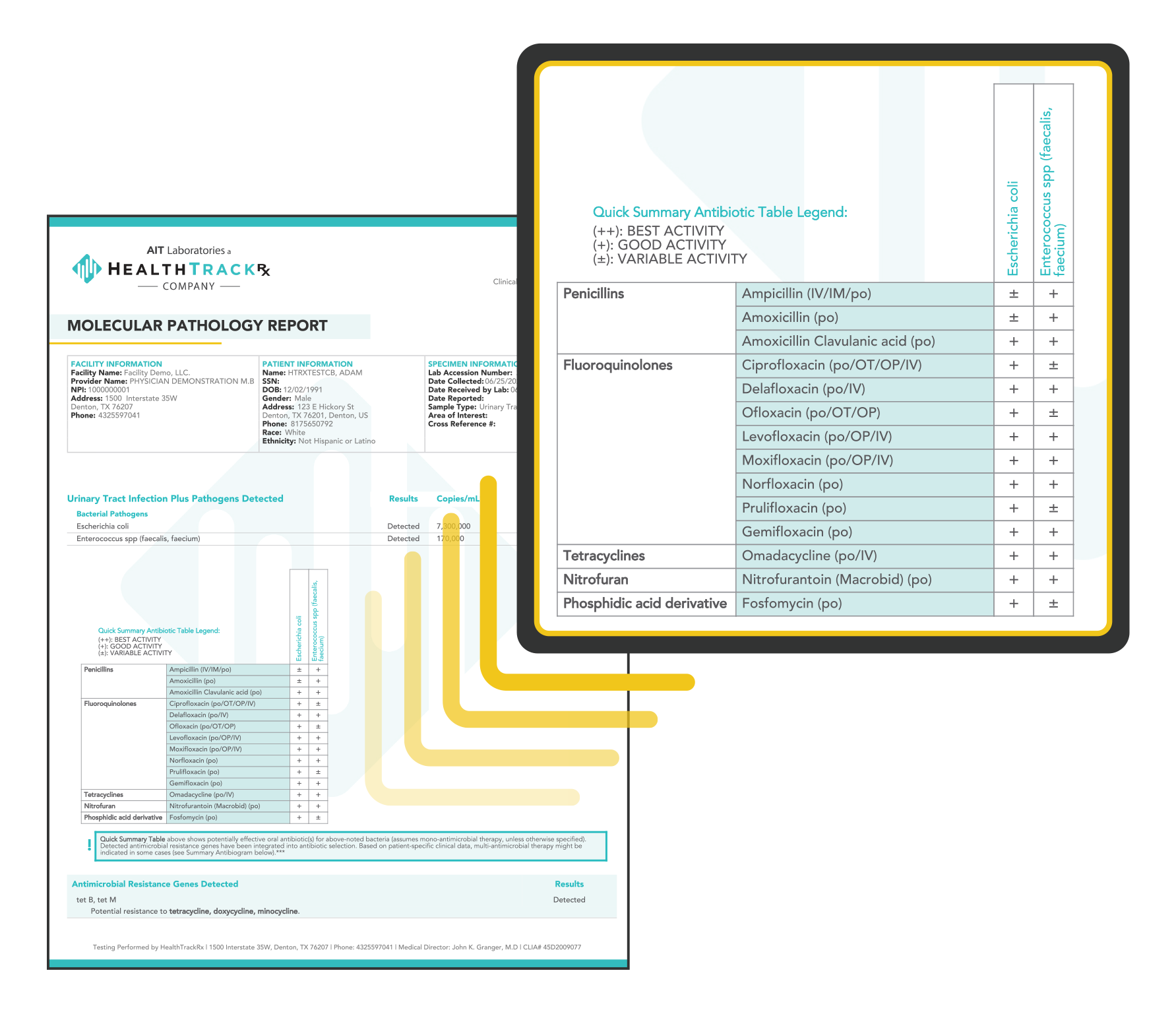
HealthTrackRx is an industry leading infectious disease laboratory providing accurate, targeted molecular diagnostics. With over 8,000,000 samples processed and next-morning results, our technological advances in PCR (Polymerase Chain Reaction) testing reshapes patient care by mobilizing faster clinical decisions and helping providers treat patients quickly and accurately. To curtail the cost of treating resistant infections repeatedly, HealthTrackRx provides a personalized, proprietary antibiogram that provides antibiotic resistance gene detection, and lists potential antibiotic efficacy.
How HealthTrackRx PCR Compares to Traditional Culture
When an accurate diagnosis matters, speed of pathogen identification and clinical actionability are key. HealthTrackRx’s PCR testing identifies what culture misses, particularly the fastidious organisms and pathogens that are especially difficult, if not impossible, to culture.
HealthTrackRx PCR Test |
vs. |
Culture Test |
|---|---|---|
| Next-Morning Results | Turnaround Time | Results can take an average of 3-14 days |
| Our pathogen assay incorporates the latest in RT-PCR technology, targeting the most relevant viral, bacterial, and fungal pathogens | Accuracy | Typically, only detects the primary pathogen |
| Rapid detection of fastidious organisms such as fungi, anaerobes, and resistance genes | Detection | Extended incubation time for fungi and anaerobes, potential for “mixed flora” and no specificity of pathogens |
| We offer ONE universal collection device | Collection | Multiple collection devices, potential for contamination |
| Less susceptible to antibiotic use | Antibiotics | Is affected by antibiotic use |
How HealthTrackRx PCR Compares to Traditional Culture
When an accurate diagnosis matters, speed of pathogen identification and clinical actionability are key. HealthTrackRx’s PCR testing identifies what culture misses, particularly the fastidious organisms and pathogens that are especially difficult, if not impossible, to culture.
Diagnostic Innovation
Syndromic Menus
Benefits of HealthTrackRx PCR
Urinary
PCR is able to detect bacteria in a higher percentage of patients that are symptomatic compared to urine culture. In addition, PCR can detect more polymicrobial infections compared to urine culture. Culture-based testing alone can miss up to 67% of UTI infections. With the rise in antimicrobial resistance, PCR testing can identify resistance genes within 24 hours, such as the resistant gene NDM-1 in E. coli and Klebsiella, leading to early effective treatment.
Urinary
Gastrointestinal
STD/STI
Ophthalmology
Our Labs
Clarksville, IN • Denton, TX • Marietta, GA • Sherman Oaks, CA
Contact Us
Our Team is Available Now
1 https://www.idsociety.org/public-health/antimicrobial-resistance/archive-antimicrobial-resistance/facts-about-antibiotic-resistance/
2 Spellberg B et al. Combating Antimicrobial Resistance:Policy Recommendations to Save Lives. Clinical Infectious Diseases 2011;52(S5):S397–S428)
3 https://amr-review.org/Publications.html
4 https://www.cdc.gov/nchs/fastats/infectious-disease.htm
5 https://www.idsociety.org/public-health/antimicrobial-resistance/antimicrobial-resistance/